A recent editorial highlighted three common low-risk CHEK2 mutations (p.I157T, p.S428F, and p.T476M) that lead to a breast cancer risk of <1.4 fold (compared to “typical” CHEK2 mutations where the risk is over 2-fold).1 This is important because the level of risk for these mutations does not warrant high-risk screening. Another study on these three low-risk mutations showed how combinations of low- and regular-risk CHEK2 mutations may affect breast cancer risk.2 Results showed differing risks for various combinations of variants as shown below.
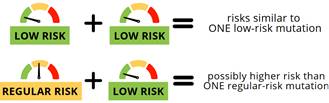
An accompanying editorial highlights the importance of contextualizing cancer risks for these low-risk variants based on the second allele (‘mutation’).3 While biallelism (meaning having two mutations) does not occur that often, this work shows the importance of discussion at the VARIANT alleles level to guide cancer risk and care.
1Hernandez et al. Oncotarget. 2024:15:459-460. PMID: 38985133. Article available at: https://tinyurl.com/fullarticle28604. Social media post available at: https://tinyurl.com/post10312024; 2Bychkovsky, et al. JAMA Netw Open. 2025;8(1):e2451361. PMID: 39745704. Article available at: https://tinyurl.com/fullarticle2828642; Social media post available at: https://tinyurl.com/post2102025; 3Rajagopal. JAMA Netw Open. 2025;8(1):e2451303. PMID: 39745708. Article available at: https://tinyurl.com/fullarticle2828646. Social media post available at: https://tinyurl.com/post2102025.
