
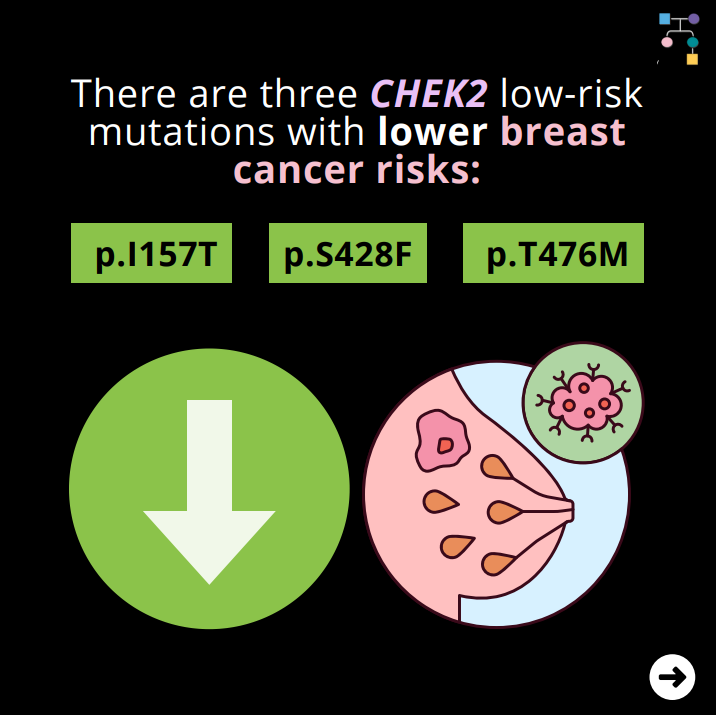


There are three CHEK2 “low-risk” mutations with lower breast cancer risks: p.I157T, p.S428F, and p.T476M.
A new study was conducted on how combinations of low- and regular-risk CHEK2 mutations may affect breast cancer risk. Results showed the following risks for various combinations of variants:
- TWO low risk = risks similar to ONE low-risk mutation
- ONE regular risk + ONE low risk = possibly higher risk than just ONE regular-risk mutation
Accompanying editorial by Dr. Rajagopal highlights:
- Findings suggest that low-risk variants are associated with a cancer risk CONTEXTUALIZED by the second allele.
- While biallelism does not occur that often, this work highlights the importance of discussion at the VARIANT alleles level to guide cancer risk and care.
Check out the articles to learn more:
⤷ https://jamanetwork.com/journals/jamanetworkopen/fullarticle/2828642
⤷ https://jamanetwork.com/journals/jamanetworkopen/fullarticle/2828646
Reference: Bychkovsky, et al. JAMA Netw Open. 2025;8(1):e2451361. PMID: 39745704.
Reference: Rajagopal. JAMA Netw Open. 2025;8(1):e2451303. PMID: 39745708.
